Our final goal is to determine the modulators of the stress response in health and disease.
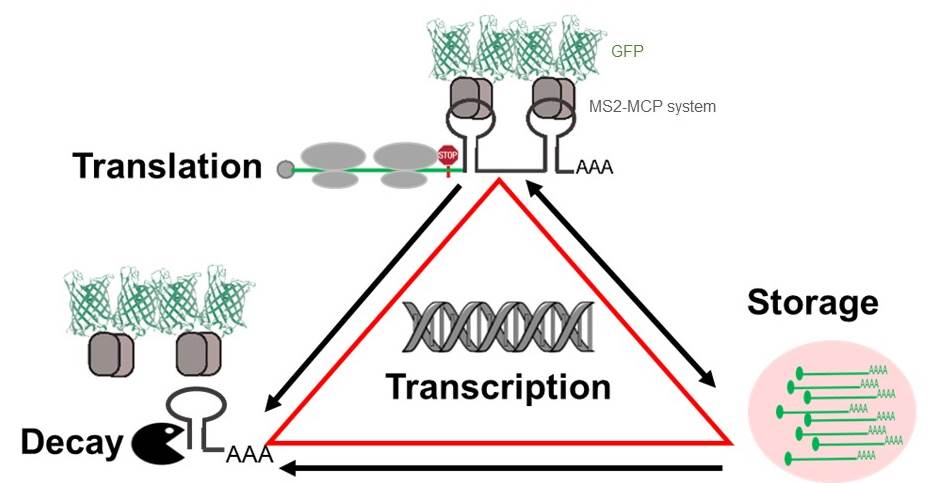
We propose that the HSP mRNAs are co-transcriptionally equipped with regulators that determine their fate in the cytoplasm (Vera et al, eLife 2014). We aim to determine how the HSP mRNAs are capable to integrate their translation, localization, and decay with the status of the cell. To this aim, we combine molecular, genetic and biochemical approaches with single-molecule fluorescence technologies to image single mRNAs and proteins in several model organisms.
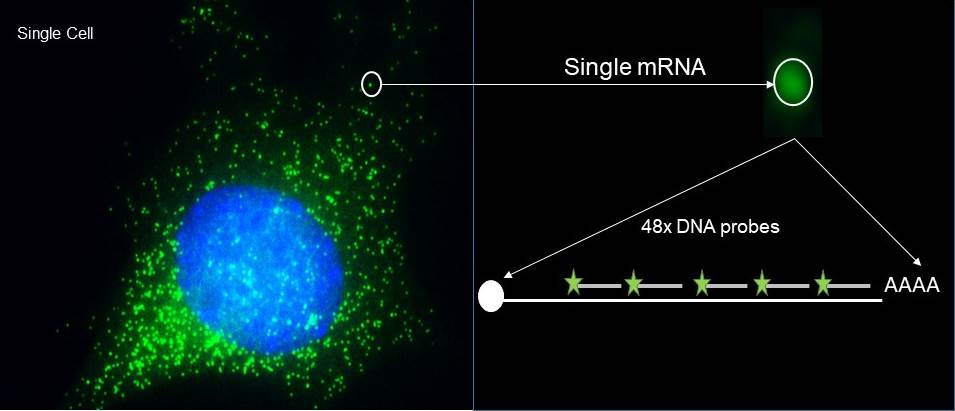
Single molecule imaging of endogenous mRNAs in fixed cells. smFISH affords visualizing up to 90% of the transcripts from a specific gene in the cell. The single molecule sensitivity relays on the high signal to noise ratio obtained from confining several fluorescent labelled DNA antisense oligonucleotides in an mRNA. This fluorescent signal is captured as a discrete spot when acquiring z-stacks in a wide field microscope equipped with a digital camera. The 3D images are then analyzed with the program FISH-quant to obtain accurate information on the brightness and localization of each mRNA molecule in the cell. These two parameters, intensity and registration, are sufficient to classify localization and measure events (Vera el al., ARG, 2016).
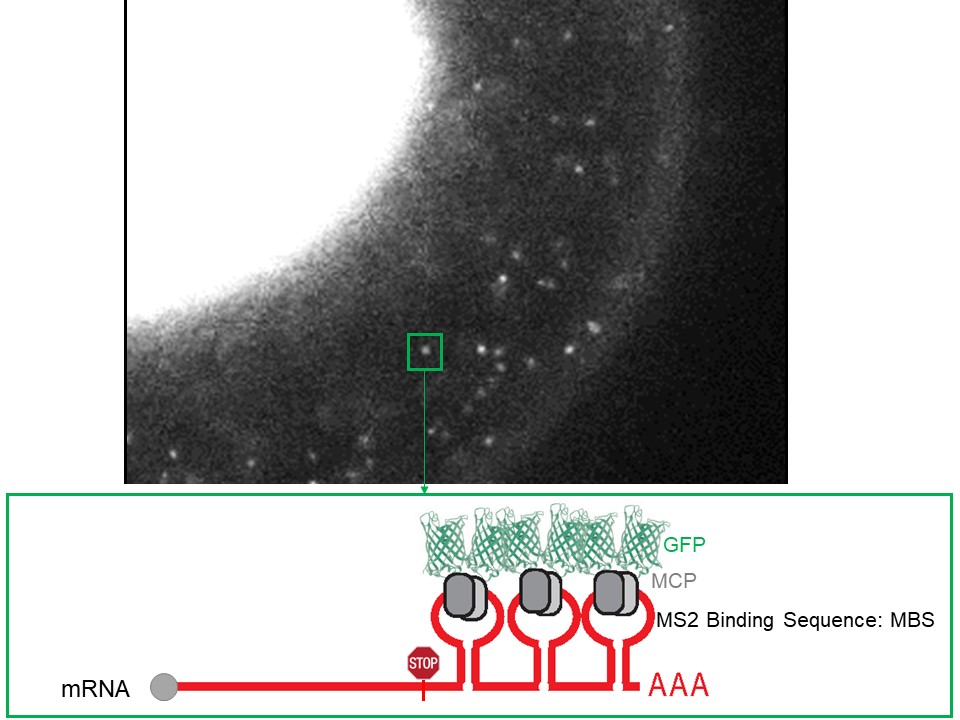
Single molecule imaging of mRNAs in live cells. We have optimized the detection of single mRNA molecules in living cells by engineering an mRNA tagging system that reports on their localization from transcription to decay. The first version of the system, known as the MS2 system, was developed in the laboratory of Dr. Singer to visualize and quantify mRNAs. Due to central role of mRNA in gene expression, these quantifications have provided unprecedented insights on the regulation of transcription, mRNA export and localization, and more recently translation. The MS2 is an RNA aptamer that is integrated within the 3’ UTR of an mRNA as an array of 24 stem loops. Each of the stem loops is tightly bound by a protein, MCP, fused to two fluorescent proteins (FP). The MS2-MCP-FP binding renders a single mRNA visible because it is decorated with 48 fluorescent molecules (Tuttuci*&Vera* et al, Nat Methods, 2018 and Vera et al, MiMB, 2019).
Single molecule imaging of translation. We visualize single peptides using the SunTag system developed by Marvin Tanenbaum (A protein tagging system for signal amplification in gene transcription and fluorescence imaging, Cell:159:635-646).